Gastrointestinal infection (GI) - February 15, 2022
Evaluating the diagnostic methods for acute gastroenteritis
Acute gastroenteritis is one of the most common causes of morbidity and mortality worldwide.1 The highest prevalence is seen in non-industrialised countries, as diseases are most frequently transmitted in poor hygienic conditions or by consuming contaminated food or water.2 However, acute gastroenteritis is still common in both low- and high-income countries.3 Symptoms and complications of acute gastroenteritis have led to a considerable disease burden due to growing health system use and productivity loss.3 For example, in the Netherlands and Belgium, annual direct medical costs of acute gastroenteritis amount to €945 million and €112 million respectively.3,4 Similarly, in Finland, productivity loss and the associated cost of lost workdays has been estimated at €1.8–2.1 million with the prevalence of participants on sick leave recorded at 3.54 times higher in the week following an outbreak.5
Acute gastroenteritis is responsible for 1.45 million deaths worldwide
Gastroenteritis is a transient disorder and can be caused by a multitude of different viral, bacterial, and parasitic pathogens.6 Norovirus is the most common pathogen responsible for acute gastroenteritis in the United States followed by Salmonella spp., Shigella spp., and Shiga toxin-producing E. coli (Figure 1).
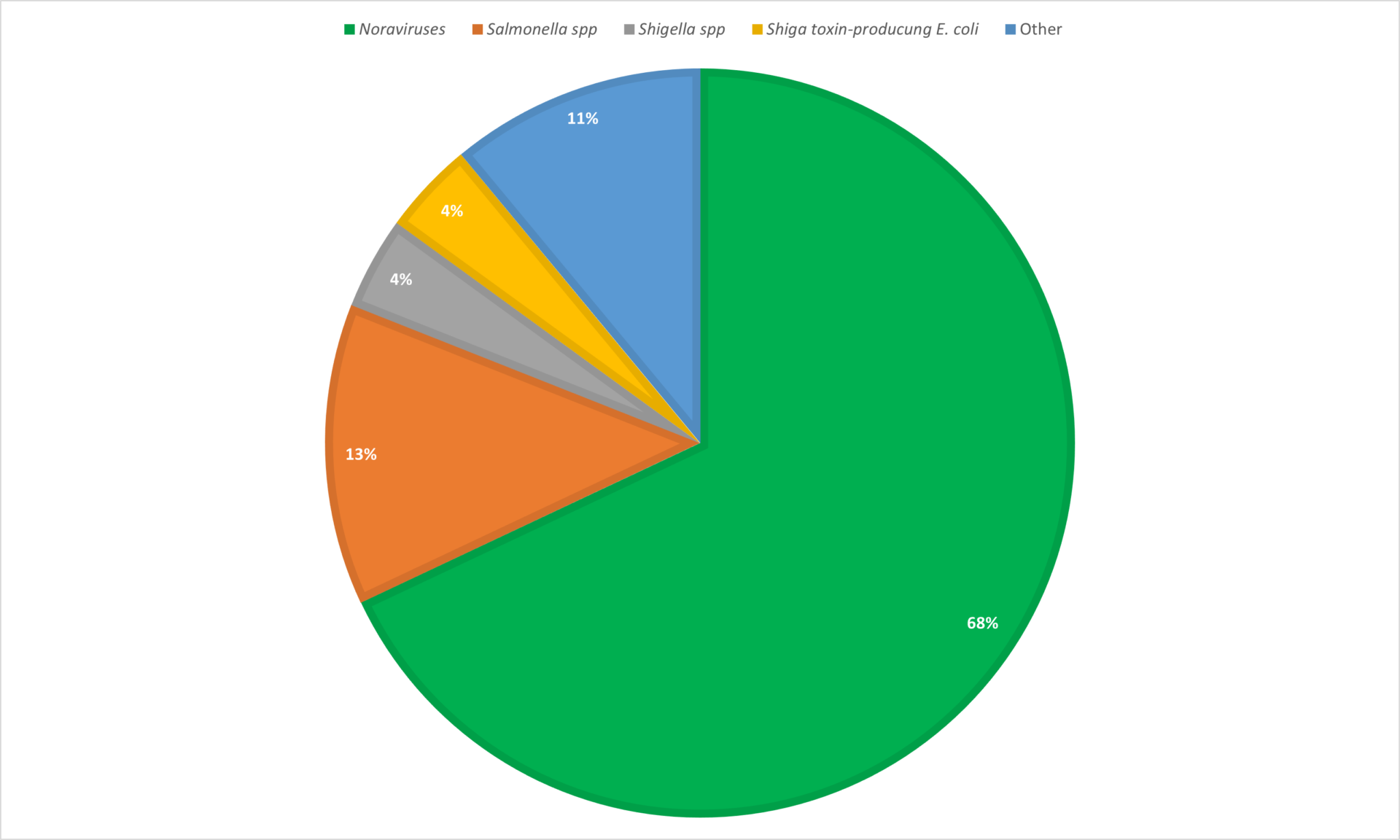
Although there has been a reduction in bacterial and parasitic infections achieved via improvements in sanitation,1 the increases in diarrhoea-related hospitalisations attributed to viral acute gastroenteritis have reached concerning levels with about 43% of childhood diarrhoea samples from developing countries containing at least one viral agent.7
Acute gastroenteritis is characterised by the sudden onset of diarrhoea, presenting with or without vomiting.6 Acute gastroenteritis is typically a self-limiting disease and most healthy individuals recover within a few days. However, complications can prove fatal for very young or very old patients.6 Complications include, but are not limited to, dehydration and electrolyte disturbance, acute kidney injury, sepsis, and haemolytic uraemic syndrome.6 Diarrhoea is the second leading cause of death in children younger than 5 years of age worldwide.8 There are 1.7 billion cases of diarrhoea and 1.45 million deaths as a result of acute gastroenteritis recorded globally every year.1,2
Rapid testing is vital
As infants and the elderly are particularly susceptible to complications arising from acute gastroenteritis, rapid provision of test results helps guide medical professionals to the appropriate treatment type for critical care patients.9 Furthermore, given the highly infectious nature of the disease, hospitals find themselves short of isolation facilities. Beds are limited and are in over demand for non-infectious patients, let alone for patients requiring isolation to prevent the spread of gastroenteritis.10
Rapid testing for gastrointestinal infections may reduce the time spent in isolation for some patients, therefore reducing overall associated burden. Several diagnostic techniques are currently in use aiming to improve patient management and avoid deadly consequences of acute gastroenteritis. In the next section, we will review them and highlight their key benefits
Conventional diagnostic techniques vs molecular testing
Conventional diagnoses of enteric gastrointestinal infections have relied on three main techniques: microscopy examination, culture and antibiotic susceptibility testing, and antigen detection using immunoassays.1,2 These methods involve several stages and procedures so that results can take up to 3 days to become available.11 For vulnerable patients, such as infants, health can decline rapidly in the face of infection, so, when time is a factor of determining survival, speed of diagnostic techniques is imperative. Each conventional diagnostic technique has advantages and disadvantages in a clinical setting.
Sensitivity disparities for parasites and virus detection
Microscopic examination remains the standard for laboratory confirmation of parasitic infections .12 Slide microscopy enables skilled technicians to visibly count the number of parasites and enables quick monitoring of the effectiveness of a given treatment.12
Although microscopy has been credited as providing rapid results, this refers to handling one sample at a time. This becomes a problem on a larger scale in a clinical setting when multiple samples are in question. Diagnostic accuracy also is dependent on the quality of the smear and equipment and ability of the microscopist.12 Molecular diagnostic platforms are available that can batch up to 24 samples within 3 hours and require less than 2 minutes of hands on time per sample.2
Electron microscopy (EM) remain the reference method for the diagnosis of viral acute gastroenteritis and polyacrylamide gel electrophoresis (PAGE) was successful in detecting rotavirus infections.13 However, when tested on the detection of norovirus, EM was recorded as having 58.3%14 sensitivity which can cause false results. A sensitivity of 90% or higher reduces the risk of false results. This risk can be mitigated by using alternative techniques such as PCR which showed a sensitivity of 94.1%.14 As well as being slow and variable in accuracy, microscopy, EM and PAGE are known for being labour intensive, requiring specialised equipment and experienced laboratory technicians.2,11
Furthermore, low positive yield of pathogens is common problem using these conventional techniques. Previous studies have indicated a positive yield as low as 2.4% in stool cultures and even lower rates of 0.1% from intensive care patients.11 Even after concentration, the minimal number of viral particles present in stool samples make direct EM fairly insensitive.1
In an effort to improve the sensitivity of EM for viral acute gastroenteritis, immune EM (IEM) was introduced which relied on specific antibodies or immune sera being added to the sample to enhance the detection of the virus.1 The associated clumping that occurs in the presence of a specific antibody greatly improves diagnostic capability.1 However IEM also requires high technical expertise and is only useful for samples collected during the very early stages of the infection.1
Problems with using culture for diagnosis of acute gastroenteritis
There are more severe cases of infectious diarrhoea caused by bacteria than any other infectious aetiology, with one study identifying a bacterial origin in 87% of cases.15 There are three common bacteria that can routinely be identified in stool cultures: Salmonella, Campylobacter, and Shigella.15 However, there are some microorganisms that cannot be easily cultured in the laboratory or require specific conditions that, if not met, will result in in failure to detect the pathogen. Another major issue is concerning long diagnostic timescales. For example, detection of Shigella by culture is possible but should be avoided for primary diagnosis due to the long turnaround times of up to 5 days.2,16 Samples will need a long incubation time and many factors affect the sensitivity of the result including type and quality of specimen, the age of the patient, appropriate transport, and culture media.2
The ability for cultures to identify pathogens has been scrutinised over the years. One study compared pathogen detection using conventional methods, including culture, versus multiplex real-time PCR on 182 patients with diarrhoea. The study found that significantly more pathogens were detected using the multiplex panel (58.3%) compared with conventional stool studies (42.3%).17 In addition to this a statistically significant difference in the test reporting time, with the multiplex panel confirming diagnosis at an average of 25 h after visiting the hospital, this reduced to 4 hours during working hours.17 Either way, the reporting time was significantly shorter than when using conventional methods, which took an average of 72 h.17
Considering this data and COVID-19 testing being at the forefront of healthcare, should we anticipate molecular diagnostic testing becoming a main form of testing in the future, with laboratories having higher capacity for molecular diagnostic tests?
Advances in molecular diagnostics allow for a targeted diagnosis of acute gastroenteritis
The introduction of Nucleic Acid Amplification Tests (NAATs) bridged the gap between conventional methods of diagnosis and the higher performing molecular diagnostic (MDx) techniques. NAATs have been consistently shown to be more sensitive and specific compared to non-nucleic acid-based methods. For viral acute gastroenteritis, diagnosis can be made more quickly in the course of the infection even when there are low levels of viral shedding.1 NAATs identify the pathogen in question irrespective of culture constraints and demonstrate good reproducibility between laboratories.18
MDx testing in symptomatic individuals has become increasingly sophisticated over the past 20 years and these diagnostic tools for infectious acute gastroenteritis have undergone several innovations from scatological cultivation to next-generation sequencing (NGS).11 NGS is characterised by the ability to sequence mixed DNA or RNA genomes rapidly and intensely and has already had a substantial impact on our understanding of the epidemiology of many diarrhoea-associated pathogens.
Commercially available MDx based assays have changed the way laboratory diagnosis of enteric infections are performed.2 Although reagent and instrument costs are higher for real-time PCR technology compared to conventional techniques, the level of automation that comes with this technology significantly reduces hands-on time per specimen to as little as two minutes.2 The automated efficiency makes this kind of targeted MDx far less labour intensive compared to traditional techniques.
PCR tests have been shown to consistently result in high diagnostic values with >90% sensitivity and close to 100% specificity.11 Multiplex PCR-based testing simultaneously evaluates stool specimens for the presence of multiple pathogens, whereas single PCR-based platforms target specific pathogens.11 Highly multiplex panels do run the risk of provided false-positive results in the case of rare pathogens.19 In addition, it is important that these platforms are integrated and do not require separate nucleic acid extraction and post-PCR handling in order to prevent contamination and false positives.2 When these criteria are met, a targeted syndromic approach can be used to test several pathogens known to cause the same syndrome in patients.
Compared to single PCR-based platforms, integrated multiplex platforms have a significant effect on patient management.20 Multiplex platforms have the potential to identify pathogens faster, allow early initiation of targeted antibiotics, alter management of antimicrobials, and thus optimise infection control.20
References
- Sidoti F, Rittà M, Costa C, et al. Diagnosis of viral gastroenteritis : limits and potential of currently available procedures. J Infect Dev Ctries 2015;9:551-561.
- Amjad M. An Overview of the Molecular Methods in the Diagnosis of Gastrointestinal Infectious Diseases. Int J Microbiol 2020;2020:1-13. Available at: https://www.hindawi.com/journals/ijmicro/2020/8135724/.
- Papadopoulos T, Klamer S, Jacquinet S, et al. The health and economic impact of acute gastroenteritis in Belgium, 2010–2014. Epidemiol Infect 2019;147:e146. Available at: https://www.cambridge.org/core/product/identifier/S095026881900044X/type/journal_article.
- Pijnacker R, Mangen M-JJ, van den Bunt G, et al. Incidence and economic burden of community-acquired gastroenteritis in the Netherlands: Does having children in the household make a difference? Riddle MS, ed. PLoS One 2019;14:e0217347. Available at: https://dx.plos.org/10.1371/journal.pone.0217347.
- Halonen JI, Kivimäki M, Oksanen T, et al. Waterborne Outbreak of Gastroenteritis: Effects on Sick Leaves and Cost of Lost Workdays. Nizami Q, ed. PLoS One 2012;7:e33307. Available at: https://dx.plos.org/10.1371/journal.pone.0033307.
- NICE. Gastroenteritis : Summary. NICE 2020. Available at: https://cks.nice.org.uk/topics/gastroenteritis/. Accessed October 1, 2021.
- Ramani S, Kang G. Viruses causing childhood diarrhoea in the developing world. 2009:477-482.
- Farthing M, Salam MA, Lindberg G, et al. Acute Diarrhea in Adults and Children. J Clin Gastroenterol 2013;47:12-20. Available at: https://journals.lww.com/00004836-201301000-00007.
- Freeman K, Mistry H, Tsertsvadze A, et al. Multiplex tests to identify gastrointestinal bacteria, viruses and parasites in people with suspected infectious gastroenteritis: a systematic review and economic analysis. Health Technol Assess (Rockv) 2017;21:1-188. Available at: https://www.journalslibrary.nihr.ac.uk/hta/hta21230.
- Sandmann FG, Jit M, Robotham JV, et al. Burden, duration and costs of hospital bed closures due to acute gastroenteritis in England per winter, 2010/11–2015/16. J Hosp Infect 2017;97:79-85. Available at: https://linkinghub.elsevier.com/retrieve/pii/S0195670117302852.
- Shen N, Tao Y, Du B-L, et al. Molecular diagnostic practices for infectious gastroenteritis. Chin Med J (Engl) 2020;133:1485-1486. Available at: https://journals.lww.com/10.1097/CM9.0000000000000841.
- CDC. Diagnosis ( Microscopy ). Cent Dis Prev Control 2021:2021. Available at: 02/08/2021CDC – Malaria – Malaria Worldwide – How Can Malaria Cases and Deaths Be Reduced? – Diagnosis (Microscopy)https://www.cdc.gov/malaria/malaria_worldwide/reduction/dx_microscopy.html. Accessed October 1, 2021.
- Beards GM. Laboratory diagnosis of viral gastroenteritis. Eur J Clin Microbiol Infect Dis 1988;7:11-13. Available at: http://link.springer.com/10.1007/BF01962164.
- Rabenau HF, Stürmer M, Buxbaum S, et al. Laboratory diagnosis of norovirus: Which method is the best? Intervirology 2003;46:232-238.
- Sattar SBA, Singh S. Bacterial Gastroenteritis.; 2021. Available at: http://www.ncbi.nlm.nih.gov/pubmed/30020667.
- Moro DD, David MO. Infectious Gastroenteritis: Causes, Diagnosis, Treatment and Prevention. Lupine 2019;2:1-6.
- Yoo IH, Kang HM, Suh W, et al. Quality Improvements in Management of Children with Acute Diarrhea Using a Multiplex-PCR-Based Gastrointestinal Pathogen Panel. Diagnostics 2021;11:1175. Available at: https://doi.org/10.3390/diagnostics11071175.
- Rodger AG, Morris-jones S, Huggett J, et al. The role of nucleic acid amplification techniques ( NAATs ) in the diagnosis of infective endocarditis. Br J Cardiol 2010;2010:195-200.
- Pankhurst L, Macfarlane-Smith L, Buchanan J, et al. Can rapid integrated polymerase chain reaction-based diagnostics for gastrointestinal pathogens improve routine hospital infection control practice? A diagnostic study. Health Technol Assess (Rockv) 2014;18:1-167. Available at: https://www.journalslibrary.nihr.ac.uk/hta/hta18530/.
- Chang L-J, Hsiao C-J, Chen B, et al. Accuracy and comparison of two rapid multiplex PCR tests for gastroenteritis pathogens: a systematic review and meta-analysis. BMJ Open Gastroenterol 2021;8:e000553. Available at: https://bmjopengastro.bmj.com/lookup/doi/10.1136/bmjgast-2020-000553.